All published articles of this journal are available on ScienceDirect.
Genetic Diversity of Two Tilapia Species (Oreochromis Niloticus and Sarthrodon Galilaeus) Using Random Amplified Polymorphic DNA
Abstract
Introduction:
In this study, the genetic variability in eight populations of Oreochromis niloticus and seven populations of Sarotherodon galilaeus was estimated using molecular markers.
Materials and Methods:
Fish specimens were collected from eight sites representing the White Nile, Blue Nile and the River Nile. Tissue samples from gills and dorsal fin were removed from individual specimens and preserved separately in absolute ethanol prior to molecular analysis by RAPD-PCR using eight primers. DNA analysis using OPA-04, OPA-13, OPA-03, OPA-06, OPA-07, OPA-09, OPA-10 and RAPD-8 produced different bands for each.
Results and Discussion:
The total bands generated by the primers were: 17, 16, 18, 12, 12, 14, 14, and 17. They were in the range of 100 to 1020 bp. Levels of variability were estimated by the proportion of polymorphic bands obtained by each primer within a population. The range of variability was wider in O. niloticus (46.0 to 91.7) compared to S. galilaeus (56.2 to 83.3). The dendrogram obtained differentiated the populations into 22 sub-clusters. Oreochromis niloticus from Al Kalakla exhibited a high level of genetic diversity. This diversity is evident among and within the studied populations, as estimated by RAPD-PCR techniques.
Conclusion:
To promote tilapia production, the study recommended increasing genetic variation within broodstocks by crossing high similarity breeds with low similarity ones.
1. INTRODUCTION
Oreochromis spp. exhibits valuable culture characteristics such as disease resistance, increased environmental tolerances, easy reproduction, and efficient use of low-protein diets, high palatability, marketability and nutrient content [1]. Thus, they are especially well-suited for culture in developing countries due to their fast growth and short generation time, tolerance to a wide range of environmental conditions, resistance to stress and disease, ability to reproduce in captivity, and their acceptance of artificial feeds right after yolk-sac absorption [2]. However, only a few of tilapia species are commercially important, and fewer are of aquaculture significance. Oreochromis niloticus, Oreochromis aureus and various hybrids of these with O. mossambicus are regarded as the most important aquaculture candidates.
Genetic variation and diversity of fish species are valuable tools in aquaculture and fisheries management in the identification of stocks, in discrete breeding populations and in estimating stock mixtures [3]. The identification of sub-populations provides attributes for assessing a number of parameters, including genetic diversity [4]. Knowledge of fish stock genetic structure is critical for stock enhancement or supportive breeding programmes [5]. It provides an overview of genetics of the populations and individuals [6]. These data contribute to knowledge about effective population size, gene flow and mating systems, which are important in managerial decisions in aquaculture [7].
According to Blankenship and Leber [8], the observable variation present in character in a population arises due to genetic and environmental effects. Such knowledge allows the definition of geographic boundaries for monitoring post-supplementation effects on genetic effective population size and/or assessing supplementation success. The population’s movement over a wide range of habitat and/or different environmental conditions may affect the gene structure [9]. However, small populations tend to present low genetic variability, with inbreeding resulting in a reduction in the fecundity and viability. According to Dinesh et al. [3], and Carvalho [10] individuals within a population with unique genotype, have high growth rates, developmental stability, variability, fecundity and resistance to environmental stresses and diseases.
Genetic analysis of organisms at the molecular level is a very important and widely practiced scientific tool for the estimation of variability. One important PCR-based genetic analysis is Random Amplified Polymorphic DNA analysis (RAPD). In the 1980s, the development of the Polymerase Chain Reaction (PCR) dramatically simplified access to genomic information, facilitating both basic research studies and a wide variety of applications [11, 12]. The RAPD analysis has also been employed in differentiating sex chromosomes [13], genetic inheritance [14], gene mapping [15] and fish conservation [16].
RAPD fingerprinting has been used for the differentiation of species of fishes [17]. Bardakci and Skibinski [18] used it successfully to differentiate the Indian major carp Labeo rohita, L. calbasu, Catla catla and Cirrhinus mrigala. DNA-based genetic polymorphism generated by RAPD fingerprinting has been used to construct a genetic linkage map for the zebrafish, Dania rerio [19]. The findings of Hassanien et al., [20] helped in understanding the broad-scale population structuring of O. niloticus, tilapia phylogeography and the nature and extent of its biodiversity. DNA fingerprinting offers great potential in aquaculture and in fisheries as a tool for the identification of individuals and population genetics [21]. DNA fingerprinting was obtained for O. niloticus, Barbus terazona and Poecilla reticulata by Harris et al. [21]. Mamuris et al. [22] noted that RAPD exhibited a more pronounced effect on populations of Mullus surmuletus. RAPD fingerprinting has been used for the detection of DNA polymorphism in colour mutant varieties of guppy, Poecilia reticulate, and tiger barb (Barbus tetrazon) [17].
The study aimed to estimate the genetic variability among different populations of two tilapia species, Oreochromis niloticus and Sarotherodon galilaeus, using RAPD-PCR.
2. MATERIALS AND METHODS
Samples of O. niloticus and S. galilaeus were randomly collected from eight and seven sites, respectively. These were: Al kalakla (K), Jebel Aulia (J) and Gitaina (G) representing the White Nile; Wad Madani (Md), Sennar (Sn) and Ad Damazin (D) representing the Blue Nile while Shendi (S) and Al Mawrada (M) representing the River Nile. No S. galilaeus were collected from the Al Mawrada fishing site.
Four specimens from each species were randomly selected from each of the fifteen populations for use in molecular analysis. From each fish, 0.5 g tissues of fins and gills tissues were removed and prepared for DNA extraction and molecular study following Ebraheem [23].
Eight molecular primers, each containing 10 base oligonucleotides, were used for amplification genomic DNA. Primers were randomly selected on the basis of GC content (60-70%) as shown in Table 1.
Samples were amplified in the PCR premix kit (i-MAX 11) added to 1.5μl primer (10mM) and 0.5 μl templates DNA. The reaction was completed to 20 μl with sterile distilled water. PCR amplification was conducted following the procedure described by Dinesh et al. [17]. Amplification was run using a Flexigene thermal cycling machine.
2.1. Annealing Temperature and Band Ranges
The optimal annealing temperature of 36ºC and band range selection (100 to 1020) were based on preliminary runs following the protocols adopted by Ebraheem [ 23 ] and Mohamed [ 24 ] on the molecular work on tilapias in the laboratory.
| No. | Primer name | Current symbols | Sequence | GC% |
| 1 | OPA-04 | RAPD1 | AATCGGGCTG | 60 |
| 2 | OPA-13 | RAPD2 | CAGCACCCAC | 70 |
| 3 | OPA-03 | RAPD3 | AGTCAGCCAC | 70 |
| 4 | OPA-06 | RAPD4 | GGTCCCTGAC | 70 |
| 5 | OPA-07 | RAPD5 | GAAACGGGTG | 60 |
| 6 | OPA-09 | RAPD6 | GGGTAACGCC | 70 |
| 7 | OPA-10 | RAPD7 | GTGATCGCAG | 60 |
| 8 | RAPD8 | RAPD8 | CCGGGAATCG | 70 |
| Primer No. | No. of Bands in | Molecular weight in bp | ||
| O. niloticus | S. galilaeus | O. niloticus | S. galilaeus | |
| 1 | 15 | 10 | 150-1020 | 100-600 |
| 2 | 14 | 12 | 100-1000 | 100-600 |
| 3 | 15 | 12 | 100-800 | 100-600 |
| 4 | 10 | 7 | 200-1000 | 200-600 |
| 5 | 10 | 10 | 100-700 | 100-500 |
| 6 | 11 | 11 | 100-700 | 100-700 |
| 7 | 9 | 9 | 150-800 | 100-500 |
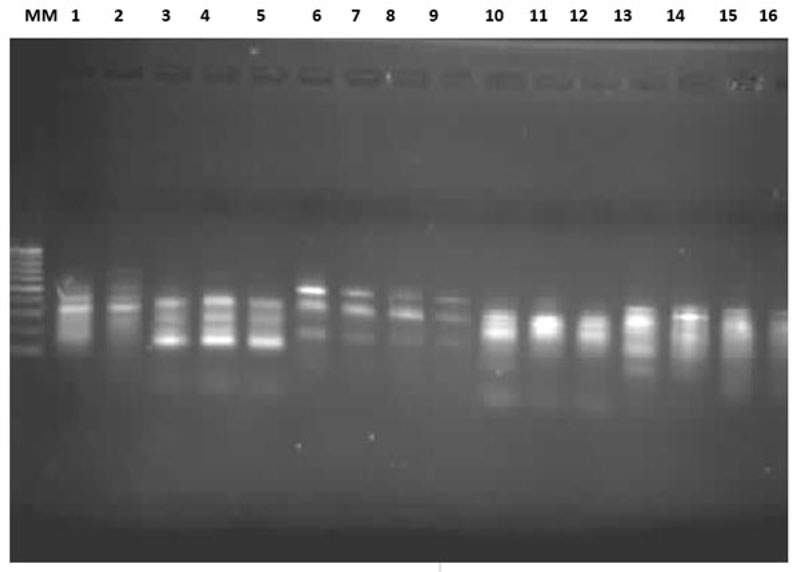
2.2. Scoring and Analysis of RAPDs
Electrophoresis was carried out as follows [23]; the gel was placed in an electrophoresis tank containing 300 ml 1X TBE running buffer at 80V for 40 min. Photographs were taken using a UVI-TECH gel documentation system. The bands were recorded on photographs as present (1) or absent (0). The number of bands generated by each primer within each population was analysed using PAST software version 3.14 [25]. The Jaccard matrix of genetic distance coefficients among each pair of population and similarity index were calculated based on pair-wise comparison between the two tilapia species.
A phenetic tree was constructed based on the Unweighted Pair Group Method with Arithmetic Mean (UPGMA) cluster analysis using PAST software version 3.14.
2.3. Data Analysis
Following Mohammed [26] and Ahmed [27], the presence (1) or absence (0) of RAPD bands were recorded; the percentage of polymorphic bands generated by each primer within each species was calculated and analyzed by Package PAST 3.14 programmer Hammer et al. [25]. The Jaccard matrix of genetic distance coefficients among each pair of population and similarity index were calculated based on pair-wise comparison between the two tilapia species. UPGMA dendrogram of population O. niloticus and S. galilaeus based on values of genetic distance calculated from data for all primers was constructed using the PAST 3.14 program provided by Hammer et al. [25].
3. RESULTS
Among the 15 O. niloticus and S. galilaeus populations, the eight primers produced bands ranging from strong, medium to sharp Distinct (Fig.1). The number of bands generated by primers 1 to 8 was: 17, 16, 18, 12, 12, 14, 14 and 17 bands, respectively. The generated bands were in the range of 100 to 1020 bp. Only the repeatable major bands ranging from 100 to 600 bp were scored for consistency. A total of 120 polymorphic bands were obtained in the 15 populations using gel electrophoresis.
The number of bands were generated by the primers was higher in O. niloticus than in S. galilaeus, except for primers 5 and 6, which gave identical band numbers. The molecular weight was higher and its range was wider in O. niloticus than in S. galilaeus (Table 2). In primer six, the molecular weight and range were similar. The general trend of differences between the number of bands and the molecular weight is due to species differences.
A different pattern of diversity was obtained in population and sites. The levels of variability in bands and % of polymorphic bands for all primers are given in Table 3. The generated bands ranged from 100 to 1020 bp. Only the repeatable major bands ranging from 100 to 600 bp were recorded for consistency. Excluding repeated bands, a total of 50 reproducible bands were obtained from 15 populations. The total polymorphic bands were 120 ranging from 64.29 to 88.23% in O. niloticus populations and from 58.33 to 83.30% in S. Galilaeus populations (Table 3). All the primers, except primer 3, detected more variability in O. nilotics than in S. galilaeus.
Jacquard matrix of genetic distance coefficients among the 15 populations of O. niloticus and S. galilaeus ranged from 0.02 to 0.27. The highest interspecies values were recorded for O. niloticus from Wad Madani, Ad Damazin, Sennar and Jebel Aulia populations. The lowest value (0.02) was obtained for S. galilaeus and O. nilotics from Al Kalakla; the same was obtained for O. nilotics from Al Kalakla and S. galilaeus from Sennar. This is indicated by a comparatively high overall interspecies pairwise divergence.
The intraspecies similarity coefficients, obtained by pair-wise comparisons of the individuals in each species, ranged from 0.35 to 0.94 and 0.42 to 0.80 for O. nilotics and S. galilaeus, respectively. The highest similarity value (0.94) was obtained for O. nilotics from Shendi and Ad Damazin. The lowest value (0.15) was from Jebel Aulia for S. galilaeus. The highest similarity value (0.80) was from Shendi and also Ad Damazin, while the lowest value (0.06) was in Gitaina individuals. The similarity coefficient represents a measure of the shared bands by two or more different populations within the same and different primers.
The distinct similarity encountered may indicate interbreeding of tilapia species, for instance, the population of S. galilaeus from Al Kalakla vs O. niloticus from Al Kalakla, also O. niloticus from Al Kalakla vs S. galilaeus from Sennar. In this respect, RAPD fingerprinting may be a useful tool for the assessment of genetic variability.
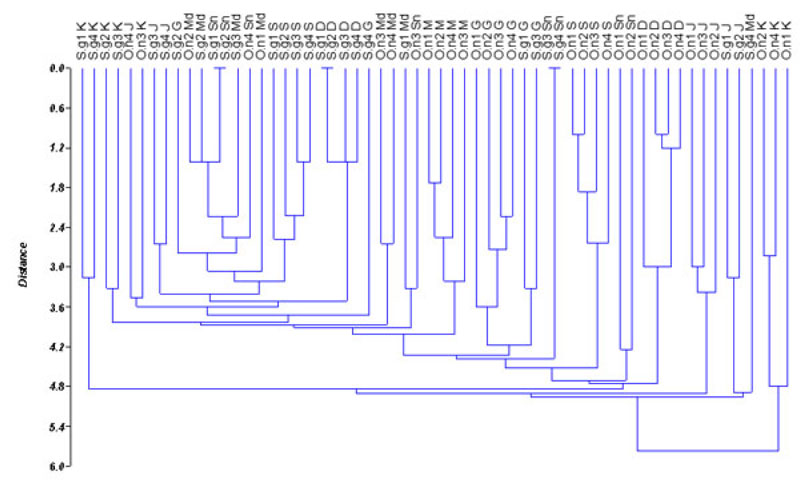
Based on the values of genetic distance, dendrogram among populations of O. niloticus and S. galilaeus is presented in Fig. (2). The dendrogram differentiates the populations into 22 sub-clusters. Sarotherodon galilaeus from Shendi, Wad Madani and some individuals of O. niloticus from Sennar were grouped together. Oreochromis niloticus from all sites except Sennar populations fell in different clusters. Similarly, S. galilaeus from all sites except Shendi populations also grouped in different clusters. Furthermore, some individuals from the same site expressed a high degree of divergence and grouped with individuals from a different site (Fig. 2). The populations of O. niloticus and S. galilaeus from Wad Madani and Sennar fell in the same cluster indicating a similarity between those populations. However, O. niloticus from Al Kalakla exhibited a high divergence level from all other tilapia populations. The obtained result indicates that the RAPD method can be used to detect variation among O. niloticus and S. galilaeus populations.
The optimal annealing temperature for the RAPD primer and DNA polymerase in this experiment was 36°C based on Ebraheem [23] and Mohamed [24]. The number of cycles kept constant through all analyses, as were denaturation, annealing and extension temperatures 37 cycles. RAPD technology is a useful tool for identifying DNA polymorphism, estimation of genetic diversity and difference of related species in fish.
|
RAPD No. |
Oreochromis niloticus | Sarotherodon galilaeus | Polymorphic Bands Total | ||||
| No. of bands | MWTbp. | Polymorphic bands% | No. of bands | MWT bp. | Polymorphic bands% | ||
| 1 | 15 | 150-1020 | 88.23 | 10 | 100-600 | 58.82 | 17 |
| 2 | 14 | 100-1000 | 87.50 | 12 | 100-600 | 75.00 | 16 |
| 3 | 15 | 100-800 | 83.33 | 12 | 100-800 | 66.67 | 18 |
| 4 | 10 | 200-1000 | 83.33 | 7 | 200-600 | 58.33 | 12 |
| 5 | 10 | 100-700 | 83.30 | 10 | 100-500 | 83.30 | 12 |
| 6 | 12 | 100-700 | 85.71 | 11 | 100-600 | 78.57 | 14 |
| 7 | 9 | 150-800 | 64.29 | 9 | 100-500 | 64.29 | 14 |
| 8 | 14 | 100-1020 | 82.35 | 10 | 100-900 | 58.82 | 17 |
4. DISCUSSION
In this work, the variation in RAPD bands generated with each primer may indicate differences in species and populations within the species. Ambak et al. [11], attributed variation in RAPD bands to the existence of one or more copies of DN per genome or variation in the annealing process. The mixed bands affect the sensitivity of PCRs results [21]. RAPD 4 and RAPD 5 show low polymorphism among the studied O. niloticus and S. galilaeus.
In O. niloticus and S. galilaeus, the sequences of RAPD fragments generated reflected a high degree of polymorphism, which may be considered as more conserved sequences. According to Soufy et al. [28], who worked on tilapias, conserved sequences are the most useful in higher taxonomic levels and evolutionary relationships. These results were also in agreement with the study of Ambak et al. [11], and Johnson et al. [19], who stated that patterns of similarities and differences between populations showed broad agreement across primers and the overall similarity level varied between primers.
The polymorphic bands among the O. niloticus populations were 64.3-91.7% and in S. galilaeus populations, they were 41.7-83.3% for RAPD primers. A high level of polymorphism is recommended for the identification of subspecies, as suggested by Wilkerson [29].
The similarity encountered may indicate interbreeding of tilapia species, for instance, the population of O. niloticus from Al Mawrada vs Ad Damazin, also O. niloticus from Al Kalakla vs S. galilaeus from Sennar. The case of possible interbreeding is between S. galilaeus from Al Kalakla vs O. niloticus from Al Kalaklaas with decreased similarity coefficient. RAPD fingerprinting is a useful tool for assessment of genetic variability and can be applied to breeding programmes in aquaculture. The reproductive program is carried out based on the similarity coefficient. Genetic variation within broodstocks with high similarity coefficient value can be increased by outcrossing with other breeds with lower similarity coefficient index Koh [30].
However, O. niloticus from Al Kalakla exhibited a high level of divergence from all other tilapia populations. High genetic variation promotes better adaptability of the populations [31]. Soufy [28] stated that very high similarity between O. niloticus and S. galilaeus increased the probability of hybridization between them. The different habitats location of a river enhances pre-existing genetic differences, providing a high inter-population structuring [32]. Abumourod, [33] studied the common patterns of genetic variations or similarities among three species of tillapine, through DNA fingerprinting analysis using RAPD PCR from EL Abbassia fish farm in Egypt, indicated the presence of many hybrids. Hybridization between closely related species improves the genetic characters and produces many strains related to the more tilapias species.
The number of cycles was kept constant through all throughout the analysis as were denaturation, annealing and extension temperatures 37 cycles. The amplification reaction ended in 10 min at 72°C. The current PCR analysis was similar to Ambak et al. [11], and Dineshet al. [17], with an increase in the number of cycles.
RAPD technology proved to be useful for the estimation of genetic diversity among different populations within each tilapia species as well as between the two studied species.
CONCLUSION
The study concluded that there is a relatively high level of polymorphism and genetic diversity within and between O. niloticus and S. galilaeus and a comparatively high overall interspecies pairwise divergence. The population of O. niloticus from Al Kalakla is quite different from other populations, and thus can be recommended for improvement of other tilapias varieties.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No humans or animals were the basis of this research.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
Not applicable.
FUNDING
The Ministry of Agriculture and Animal Wealth and Irrigation, Khartoum State, financed this work.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
Thanks are due to the fisheries administrations in the different states of Sudan for facilities.


